Examples
Running mandrake on a multiple sequence alignment:
mandrake --alignment sub5k_hiv_refs_prrt_trim.fas --kNN 500 --cpus 4 \
--maxIter 30000000
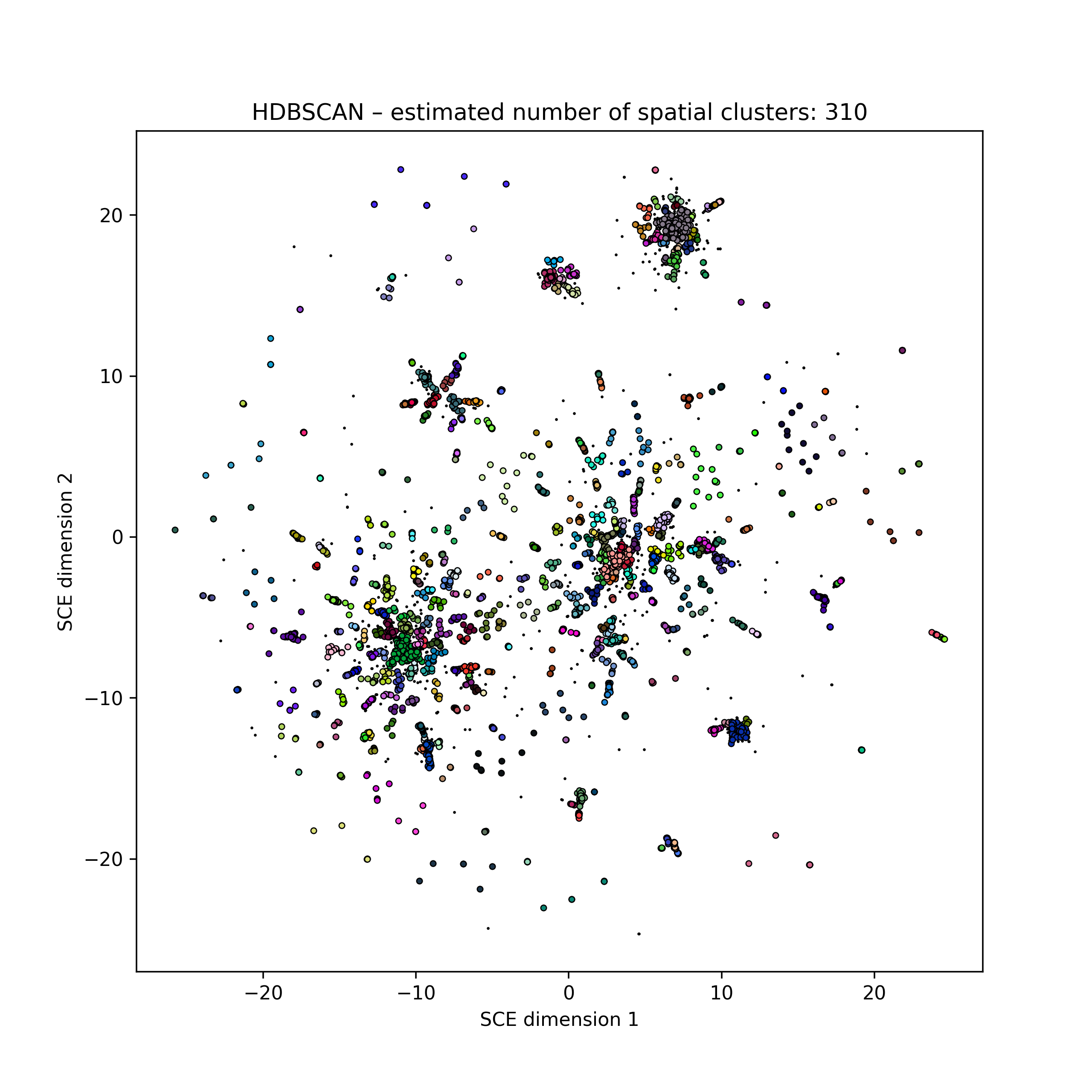
Embedding of 5k HIV pol sequences (perplexity 15)
Re-running at a different perplexities using these distances:
mandrake --output perplexity50 --distances mandrake.npz \
--perplexity 50 --cpus 4 --maxIter 30000000
mandrake --output perplexity5 --distances mandrake.npz \
--perplexity 5 --cpus 4 --maxIter 30000000
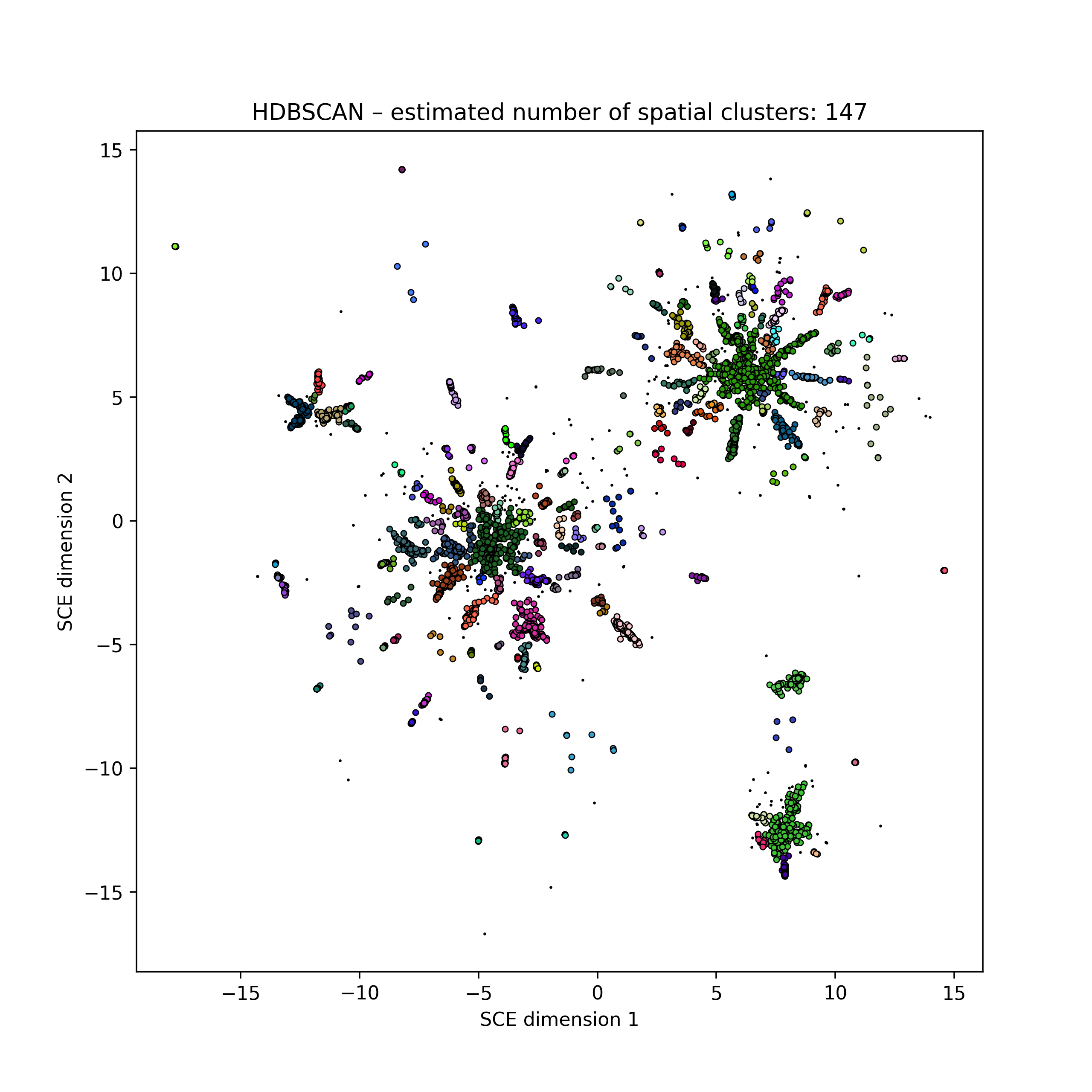
Embedding of 5k HIV pol sequences (perplexity 50)
Embedding of 5k HIV pol sequences (perplexity 5)
Running mandrake on a sketch database, and producing an animation:
mandrake --sketches mass_sketchlib.h5 --kNN 500 --cpus 4 \
--maxIter 1000000 --animate
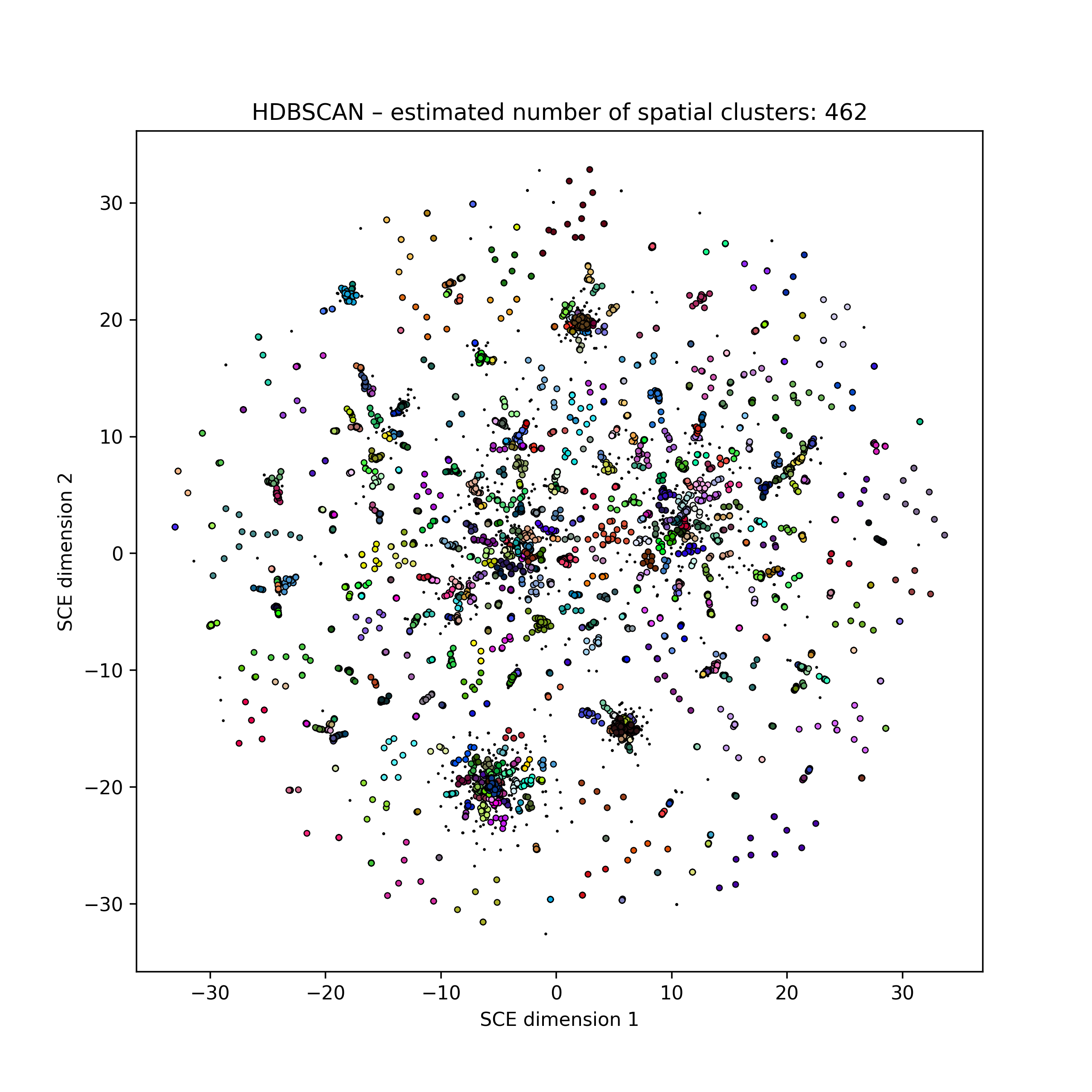
Embedding of 616 S. pneumoniae genome core distances
Running mandrake on gene presence/absence matrix from panaroo:
mandrake --accessory gene_presence_absence.Rtab --kNN 50 --cpus 16 \
--perplexity 15 --bInit 0 --maxIter 1000000000
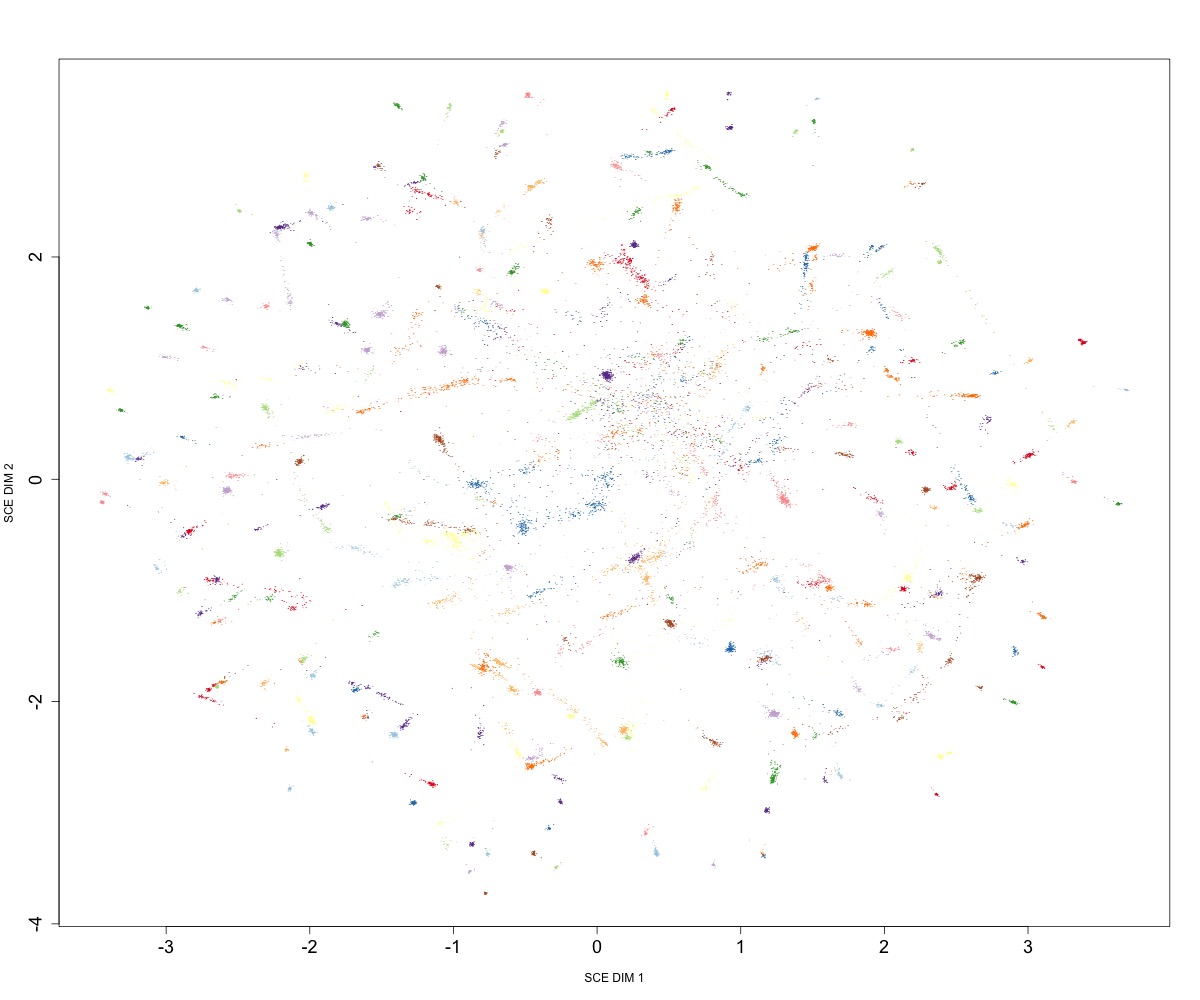
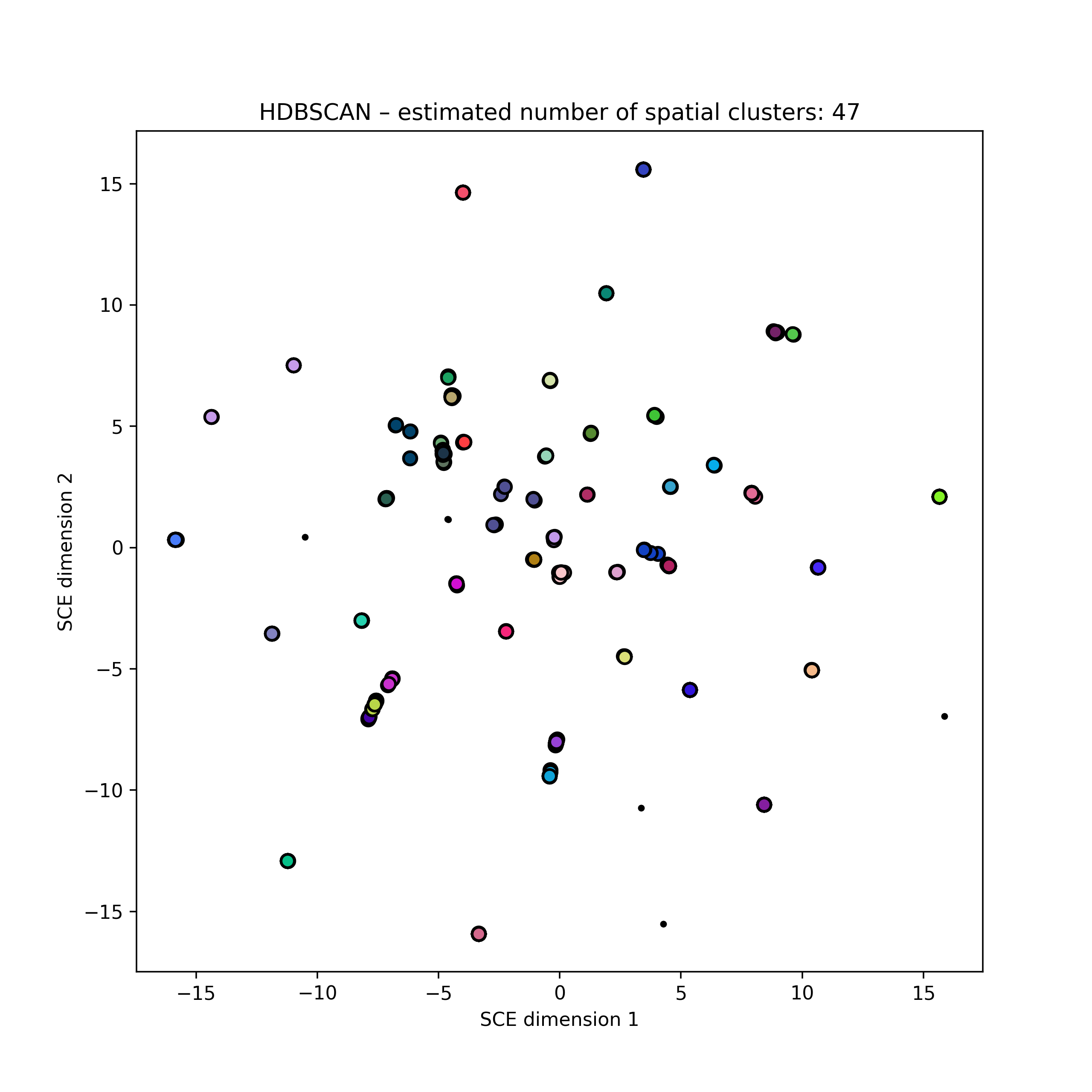
Embedding of ~20k S. pneumoniae accessory genome distances
Running mandrake on a very large multiple sequence alignment:
mandrake --cpus 60 \
--alignment SC2.fasta \
--output sc2million \
--kNN 50 --perplexity 15 --bInit 0 --maxIter 10000000000
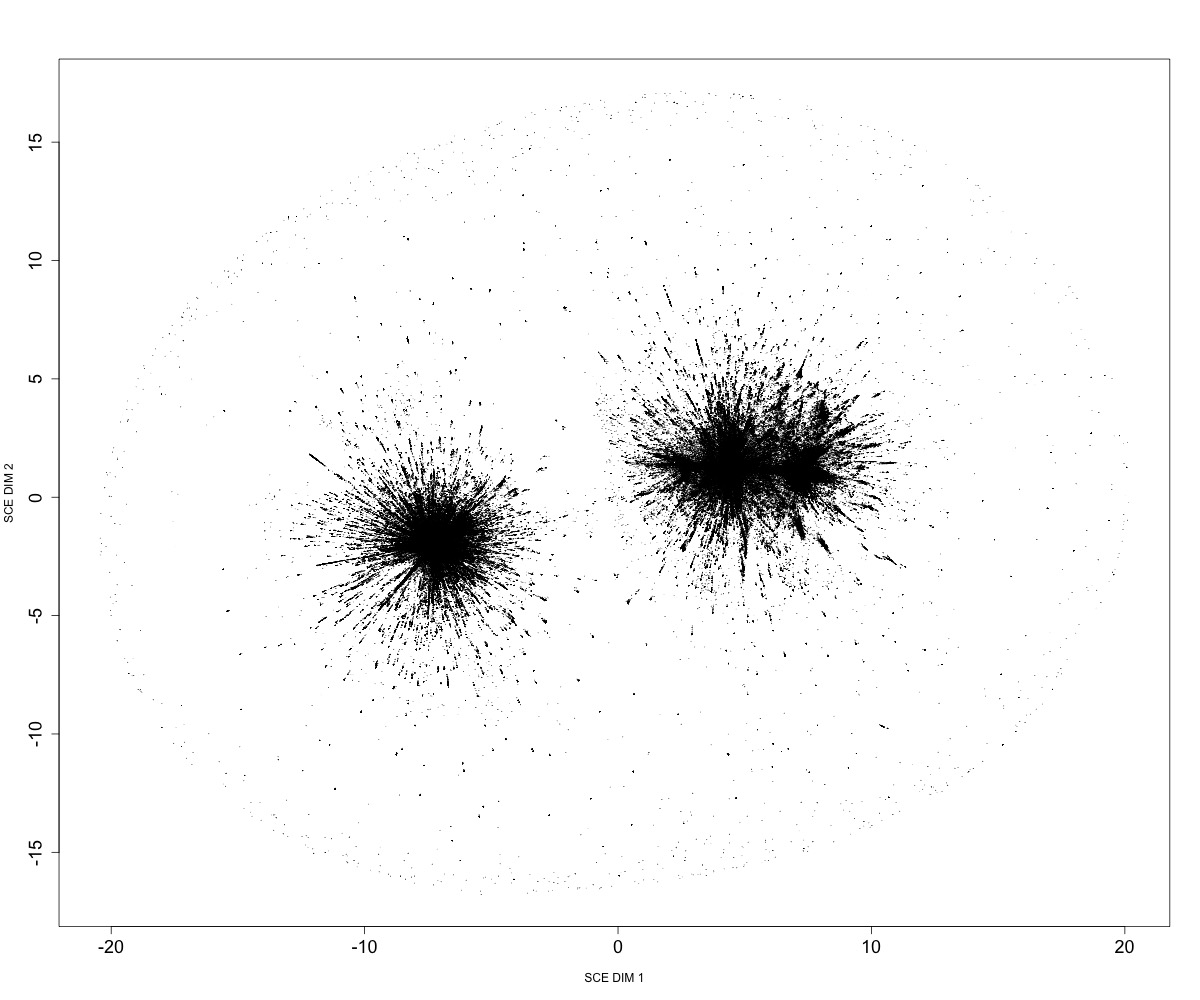
Embedding of ~1M SARS-CoV-2 genomes
Re-running mandrake using a GPU:
mandrake --cpus 60 \
--distances SC2.fasta \
--output sc2million.npz \
--perplexity 50 --no-clustering\
--n-workers 94976 --maxIter 1000000000000 --use-gpu \
--animate --labels sc2million_pangolin.txt --no-html-labels